Comparison of Artificial Neural Networks for Low-Power ECG-Classification System
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License(http://creativecommons.org/licenses/bync/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Electrocardiogram (ECG) classification has become an essential task of modern day wearable devices, and can be used to detect cardiovascular diseases. State-of-the-art Artificial Intelligence (AI)-based ECG classifiers have been designed using various artificial neural networks (ANNs). Despite their high accuracy, ANNs require significant computational resources and power. Herein, three different ANNs have been compared: multilayer perceptron (MLP), convolutional neural network (CNN), and spiking neural network (SNN) only for the ECG classification. The ANN model has been developed in Python and Theano, trained on a central processing unit (CPU) platform, and deployed on a PYNQ-Z2 FPGA board to validate the model using a Jupyter notebook. Meanwhile, the hardware accelerator is designed with Overlay, which is a hardware library on PYNQ. For classification, the MIT-BIH dataset obtained from the Physionet library is used. The resulting ANN system can accurately classify four ECG types: normal, atrial premature contraction, left bundle branch block, and premature ventricular contraction. The performance of the ECG classifier models is evaluated based on accuracy and power. Among the three AI algorithms, the SNN requires the lowest power consumption of 0.226 W on-chip, followed by MLP (1.677 W), and CNN (2.266 W). However, the highest accuracy is achieved by the CNN (95%), followed by MLP (76%) and SNN (90%).
Keywords:
ECG, ANNs- MLP, CNN, SNN, Low-Power1. INTRODUCTION
Cardiovascular diseases (CVDs) are the leading cause of death in the developed world, constituting approximately one-third of deaths [1]. To treat these deaths, long-term Electrocardiogram (ECG) monitoring is required. A cardiologist must manually examine acquired ECG signals and recognize healthy or diseased ECG i.e., recognize normal/abnormal patterns. Therefore, computer-based automated analysis is recommended for early and accurate diagnoses. Different approaches have been proposed to develop automated recognition and classification of ECG. Multilayer perceptron (MLP), convolutional neural networks (CNNs), recurrent neural networks (RNNs), long short-term memory, spiking neural network (SNN) systems, and combinations of different approaches have been proposed to improve performance and power consumption [2].
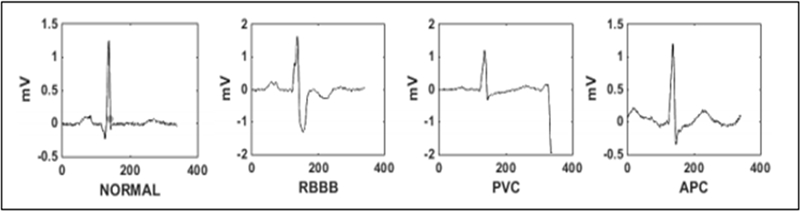
Arrhythmia is a representative type of CVD that refers to any irregular change from normal heart rhythms. Several types of arrhythmias exist, including APC, LBBB, and PVC [3], as shown in Figure 1. However, single arrhythmic beats may not affect life significantly; continuous arrhythmia beats can result in fatal circumstances. For example, prolonged premature ventricular contraction (PVCs) beats occasionally become ventricular tachycardia or ventricular fibrillation beats, which finally results in heart failure. Therefore, it is important to periodically monitor heart rhythms to manage and prevent CVDs. Additionally, resource and power requirements for extracting the most relevant information from ECG signals limit the portability of these algorithms on wearable devices. Meanwhile, machine-learning approaches allow hidden features to be obtained in the ECG signal.
Excellent heartbeat classification accuracies have been achieved by various artificial neural networks (ANNs); however, owing to its bulky operations of convolution, thresholding, pooling, and classification, which are memory and power intensive, large systems such as multicore CPUs and GPUs are required. Therefore, three different ANNs have been compared in this paper in terms of power and accuracy: multilayer perceptron (MLP), convolutional neural network (CNN), and spiking neural network (SNN) only for the ECG classification.
The remainder of this paper is organized as follows. The background of ECG, ANNs, and FPGA are discussed in Section 2. The overall design flow and experimental setup are provided in Section 3. Experimental results are presented in Section 4 and conclusions in Section 5.
2. BACKGROUND
2.1 Electrocardiogram
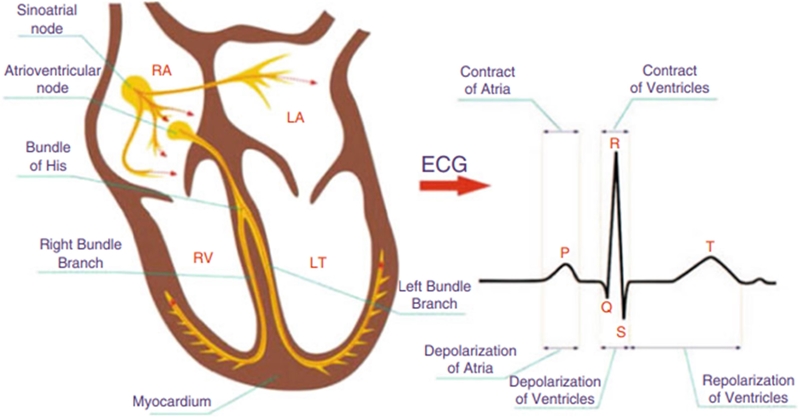
ECG is a prevalent method used in the medical field [3][4] for typical heart tests. Figure 2 shows the source of the ECG signal and a typical ECG waveform with distinct morphologies, namely P-wave, QRS complex, and T-wave.
P-wave indicates the depolarization of the left and right atrium. The wave height is less than 2.5 mm and 0.11 s in width. QRS complex starts before ventricular contraction and is the result of rapid succession. The measurement is 0.12–0.20 s and the amplitude is 5–30 mm. T-wave represents ventricular repolarization, which is after the QRS complex. It typically exhibits an amplitude of less than 10 mm in the case of limb leads.
In most previous studies, the QRS complex has been utilized to recognize arrhythmias and distinguish the strange ECG signals. QRS detection is difficult because it is subject to physiological variations owing to patients and different noise types [4]. ECG classification involves four steps: preprocessing, feature extraction, normalization, and classification. ECG preprocessing includes baseline wander, power line interference, and muscle artifact [5], which can be removed using discrete wavelet transform [6]. Feature extraction and classification is crucial. Several feature extraction methods have been applied, such as discrete Fourier transform, fast Fourier transform (FFT) [7], wavelet transform [8], higher-order statistics [9], and principal component analysis [10]. Various machine learning techniques are used as the classifier. Ubeyli [11] proposed an RNN classifier with an eigenvector-based feature extraction method and achieved 98.06% accuracy. Park et al. [12] proposed a K-NN classifier for detecting ECG arrhythmias, which resulted in 97.1% sensitivity and 98.9% specificity on average. In terms of existing algorithms, Kiranyaz et al. [13] introduced a one-dimensional CNN for ECG arrhythmia classification. Moreover, the 2D-CNN developed by Tae Joon Jun et al. [14] yielded an accuracy of 99.05%.
2.2 ANNs
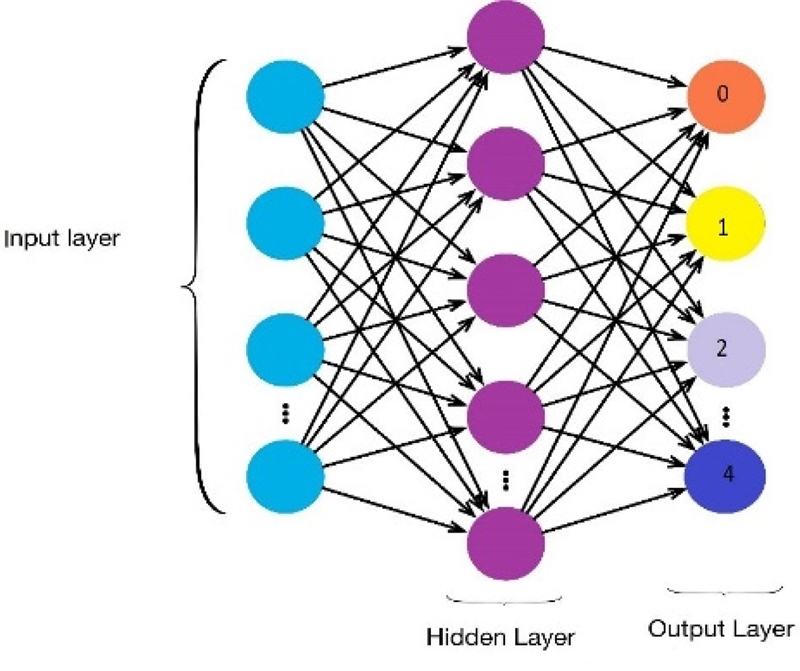
Among the most widely used network topologies from ANNs, MLP is considered as the pioneer for classification purposes [15]. Figure 3 shows an MLP classifier with an input, hidden, and output layer for classifying four different arrhythmias.
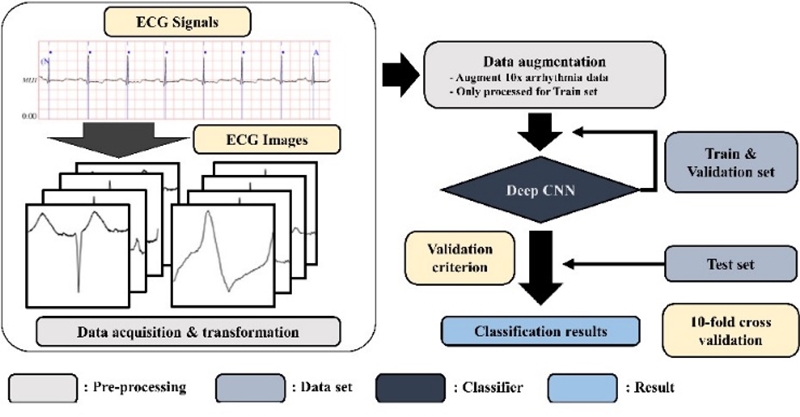
Another popular neural network is the CNN, which is a supervised method used for classification. It comprises an input and an output layer as well as multiple hidden layers. These layers are generally categorized into three types: CONV, POOL, and fully connected. In this paper, a 2D CNN was applied, which is illustrated in Figure 4. The CNN model handles two-dimensional image as an input data, ECG signals are transformed into ECG images during the pre-processing. Resultant images are augmented in order to improve the classification accuracy. The training and validation set is allotted by 80% and 20 % of the whole dataset respectively. In addition, the classification is performed on the ECG waveform images.
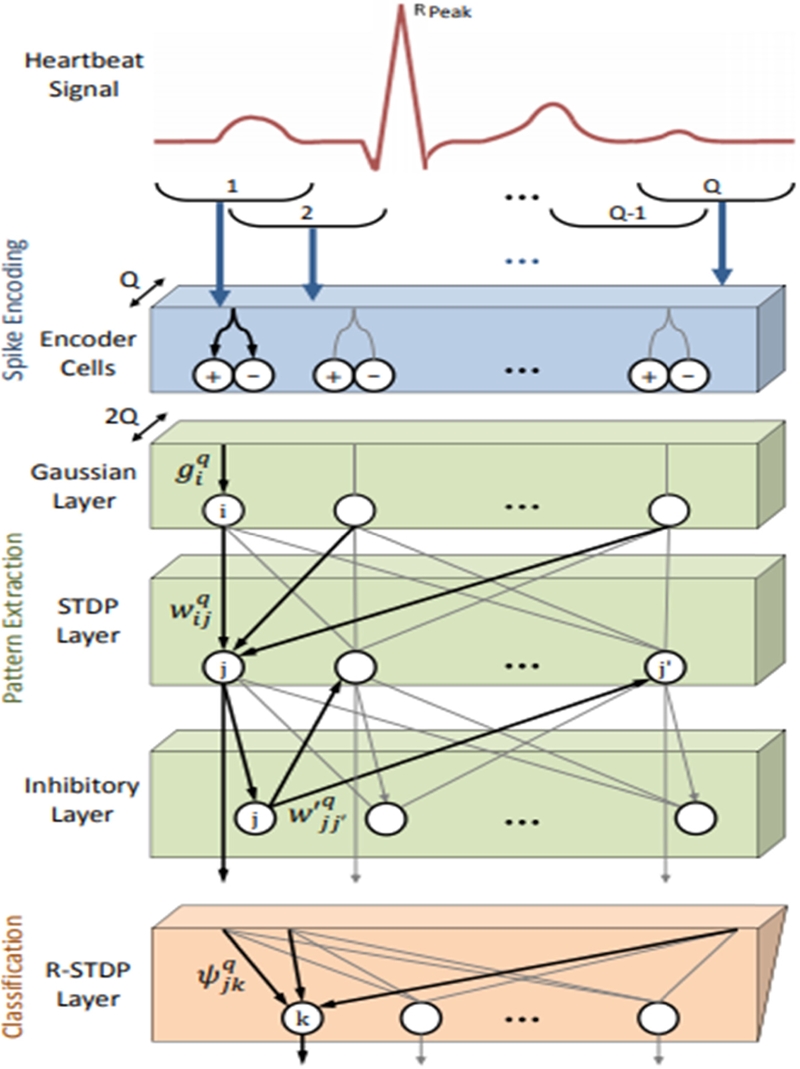
Recently, SNNs have been used to solve classification problems as an alternative to CNNs. SNNs are bio-inspired, event-driven, analog neural networks that, when mapped to dedicated neuromorphic hardware such as Loihi, CxQuad, and NeuCube, are highly energy and resource efficient compared with CNNs. This is primarily owing to a nondifferentiable discrete activation function in SNNs. Das et al., [16] proposed encoding ECG signals into a spike train (using temporal coding), and the network used was an unsupervised, liquid state machine. The approach demonstrates high classification accuracy and low power consumption for 23 distinct heartbeat classes. In this paper, a SNN architecture [17] is deployed to differentiate ECG arrhythmias. The network architecture is shown in Figure 5.
The ECG signal was split into several overlapping windows, which were subsequently encoded into spike trains. STDP represents spike-timing-dependent plasticity, where the weights are trained according to the respective timing of the spike signals. Therefore, the patterns from the spike trains are automatically extracted to the STDP layer, followed by a Gaussian layer and an inhibitory layer. Finally, an R-STDP layer was used to differentiate the extracted features. It is noteworthy that all the layers were performed in the spike domain; therefore, energy was reduced.
2.3 FPGA for Embedded AI
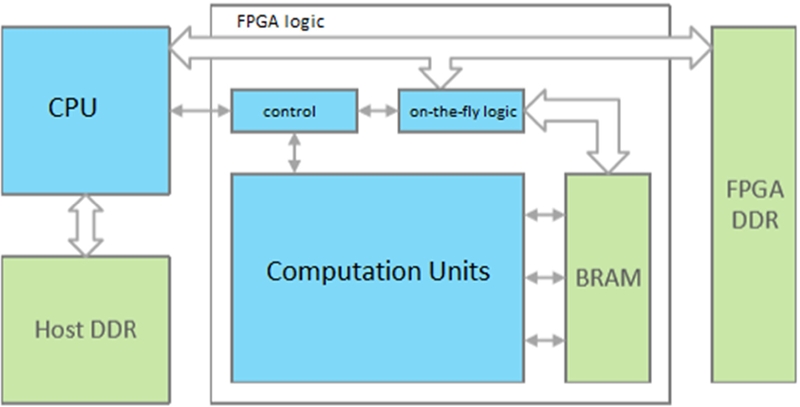
FPGAs have existed for decades. AI algorithms have been increasing in size and complexity, and GPU development has not been able to keep pace. An alternative is the FPGA, which is inherently parallel and hardware programmable. An FPGA was used in this paper owing to its flexibility, which is the most important aspect of FPGAs. The primary source of FPGAs is the configurable logic block, which uses lookup tables, flip flops, multiplexers, and switch matrices to perform a particular logic function. A typical FPGA-based neural network accelerator system is shown in Figure 6. The blue boxes denote the logic parts of the system. The workloads or commands of the FPGA logic part and the monitoring of the work status is handled by the host CPU. On the logic part of the FPGA, a controller (FSM/instruction decoder) is typically implemented to communicate with the host and generates control signals to other modules on the FPGA. The on-the-fly logic part is implemented for certain designs in case data are loaded from external memory. It can be a data arrangement module, data shifter, FFT module, etc. The green boxes represent the memory hierarchy of the system, which is categorized into three parts: host DDR, FPGA DDR, and on-chip block RAM [18].
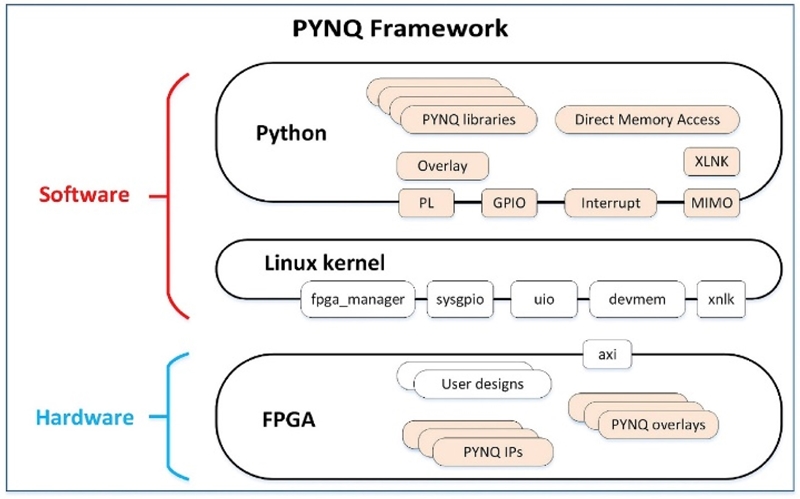
In this paper, the PYNQ-Z2 board was used, which was designed to be used with the open-source framework, as shown in Figure 7.
PYNQ denotes Python productivity for ZYNQ. From the hardware architecture perspective, the core chip of PYNQ is a Xilinx ZYNQ Chip, which is a FPGA SOC platform combining a programmable logic and programmable system (PS).
3. EXPERIMENTAL DESIGN AND SETUP
3.1 PYNQ board setup
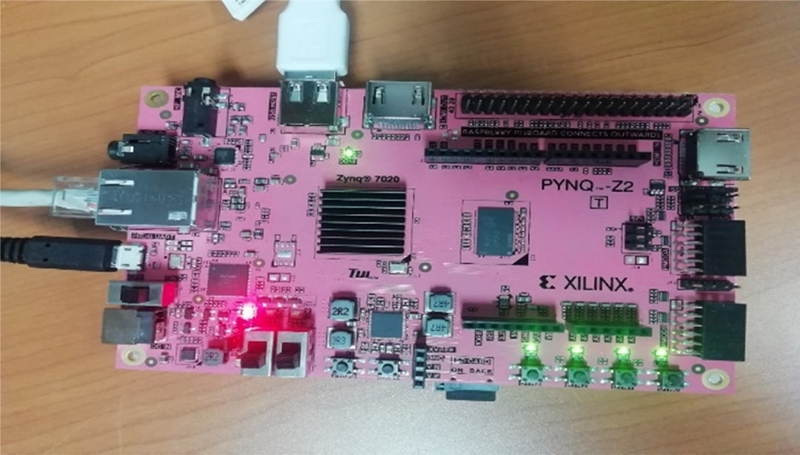
As shown in Figure 8, a red LED will illuminate immediately to indicate an ON state. Subsequently, after a few seconds, a yellow/green LED will illuminate to indicate that the Zynq device is operational.
3.2 Development Tools
Vivado IDE: An integrated development environment used for creating the hardware base system. All aspects of the system’s hardware are handled here: PS configuration, interfaces, intellectual property (IP) cores, external connections, etc.
Vivado HLS: A design tool for the synthesis of digital hardware directly from a high-level algorithm description developed in C and C++.
Jupyter notebook: A server-client application that allows notebook documents to be edited and executed via a web browser.
3.3 ECG Dataset
The data were obtained from the widely known Massachusetts Institute of Technology arrhythmia database. It contains 48 half-hour two-channel ECG recordings, obtained from 47 participants. The recordings were digitized at 360 samples per second per channel with an 11-bit resolution over a 10 mV range. In this paper, ECG heartbeat signals were converted into 2D heartbeat images for analysis using ANNs.
3.4 Hardware Design Process
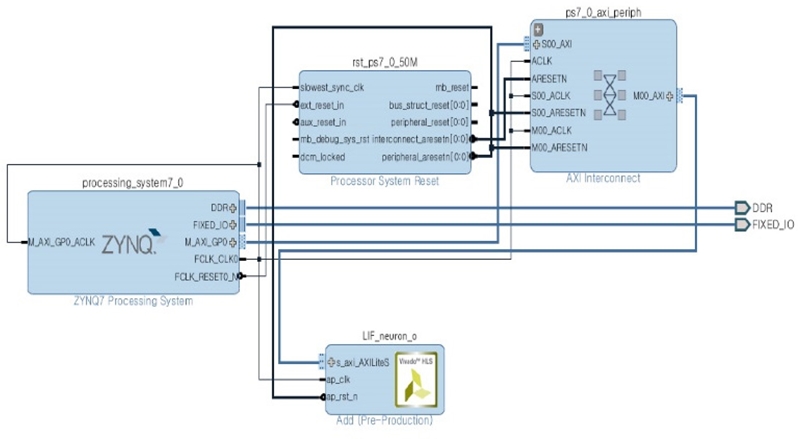
To classify the ECG data, a single hardware node is required that can interface with a software environment. Therefore, this node was created and packaged using Xilinx Vivado Design (2016.3 Edition). Figure 9 shows an example of a simple neuron for MLP with the C++ source and interface directives.
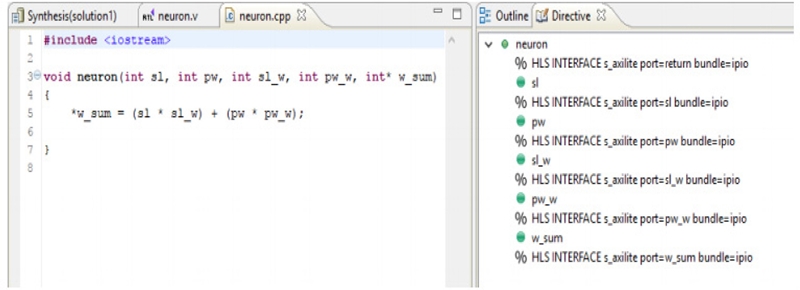
C++source and interface directives, w sum = sl(sl_w) + pw(pw w), where w_sum is the weighted sum; sl, sl_w, pw, and pw_w are the ECG signal features.
The behavior of the hardware module was specified in C++ using Vivado HLS. The RTL is ready to be packaged as an IP and exported to Vivado.
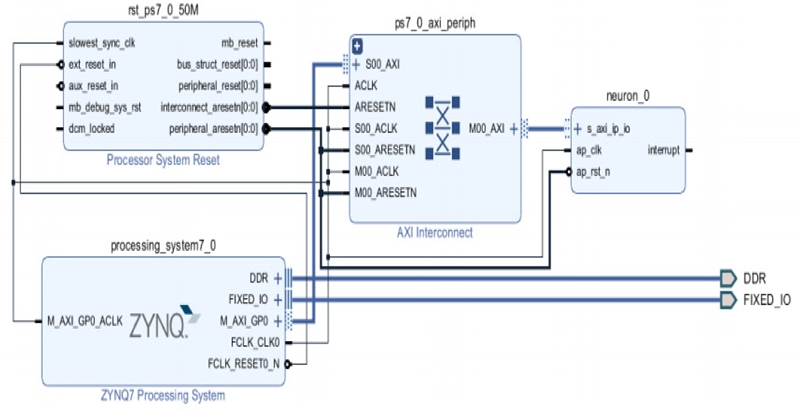
Once the IP has been exported from the HLS, it can be added to the IP integrator catalog and instantiated on the block diagram illustrated in Figure 10.
Using Figure 10, two overlay files, mlp.tcl and mlp.bit, were generated; subsequently, they were deployed into the PYNQ board. The Jupyter notebook subsequently implemented the MLP and calculated the error using Equation 1.
| (1) |
, where Yi(n) is the system response at PE i at iteration n, and di(n) is the desired response for a given input pattern of instantaneous error ei(n) .
The weight updating is expressed in Equation 2.
| (2) |
, where the local error δi(n) can be directly computed from ei(n) at the output PE or computed as a weighted sum of errors at internal PEs.
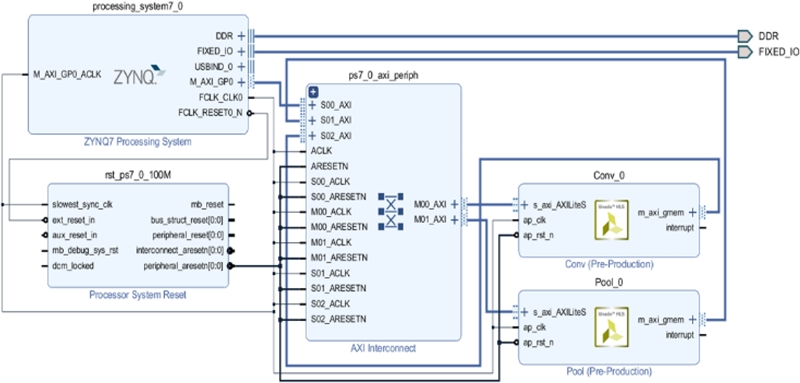
In the case of CNNs, Figure 11 shows the block of the single layer, and the convolution operation can be expressed as
| (3) |
where represents the result of the convolution layer, the weight of the convolution filter, l the number of layers, f the number of filters, the biases of the filters, fc the nonlinear activation function. Equation 4 describes the max pooling.
| (4) |
where X is obtained by implementing a maximum pool method to select features.
Neuron model used: Leaky integrate-and-fire model. The differential equation describes the membrane potential of each neuron at time t as follows:
| (5) |
, where uj(t) is the membrane potential of neuron j.
Equation 6 expresses the weight updating:
| (6) |
, where a + and a − are learning rates, and τ is the time constant. It is noteworthy that a + and a − are positive and negative constant values, respectively.
4. EXPERIMENTAL RESULTS
4.1 Accuracy analysis
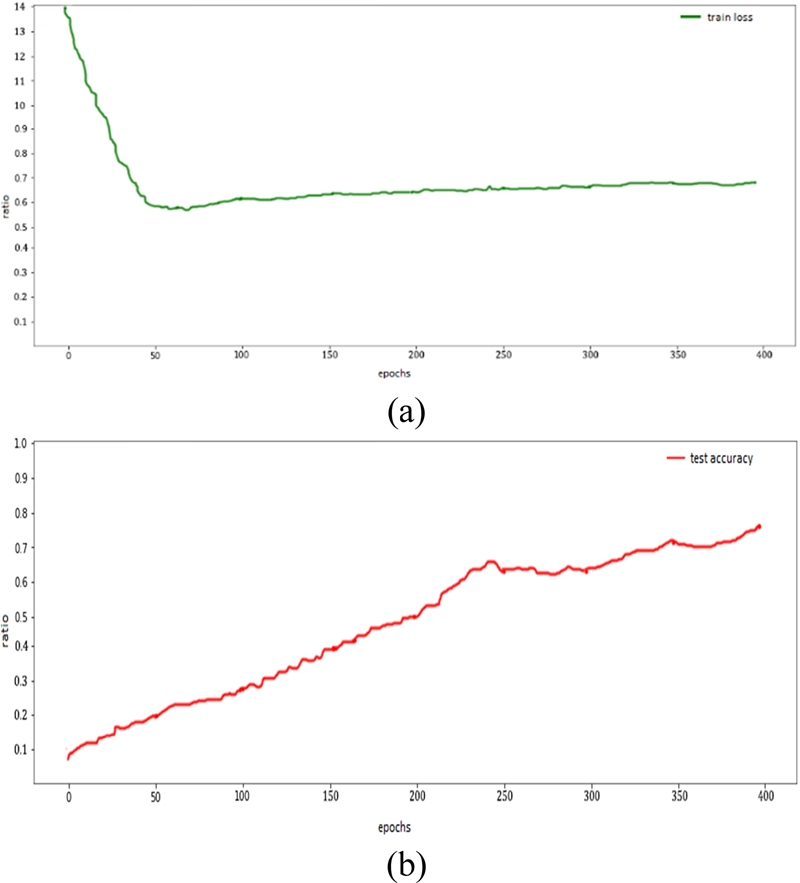
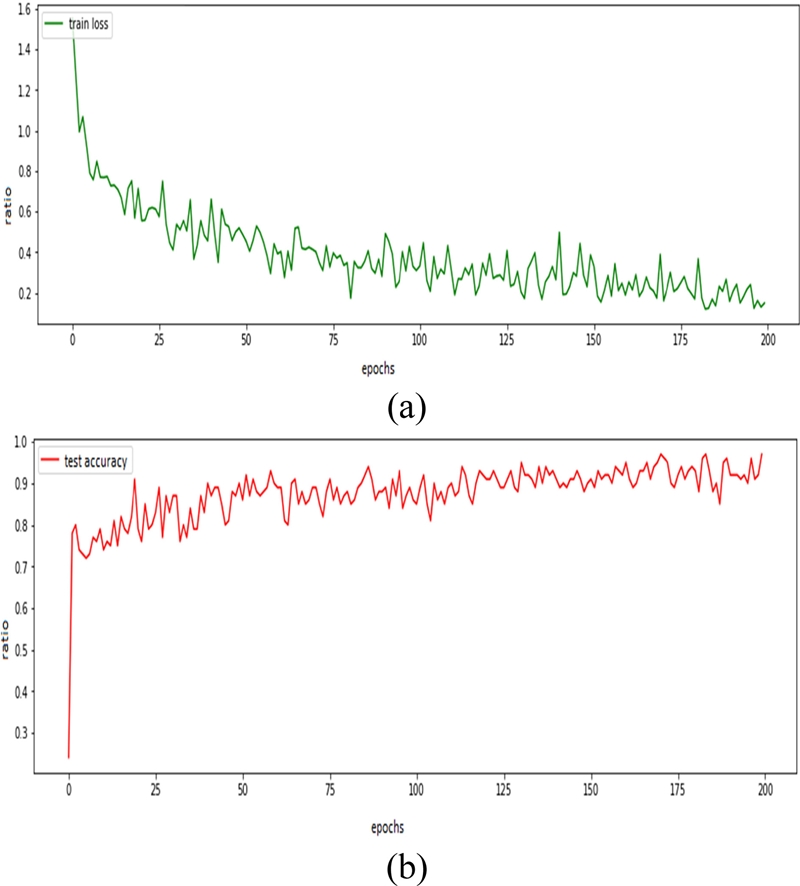
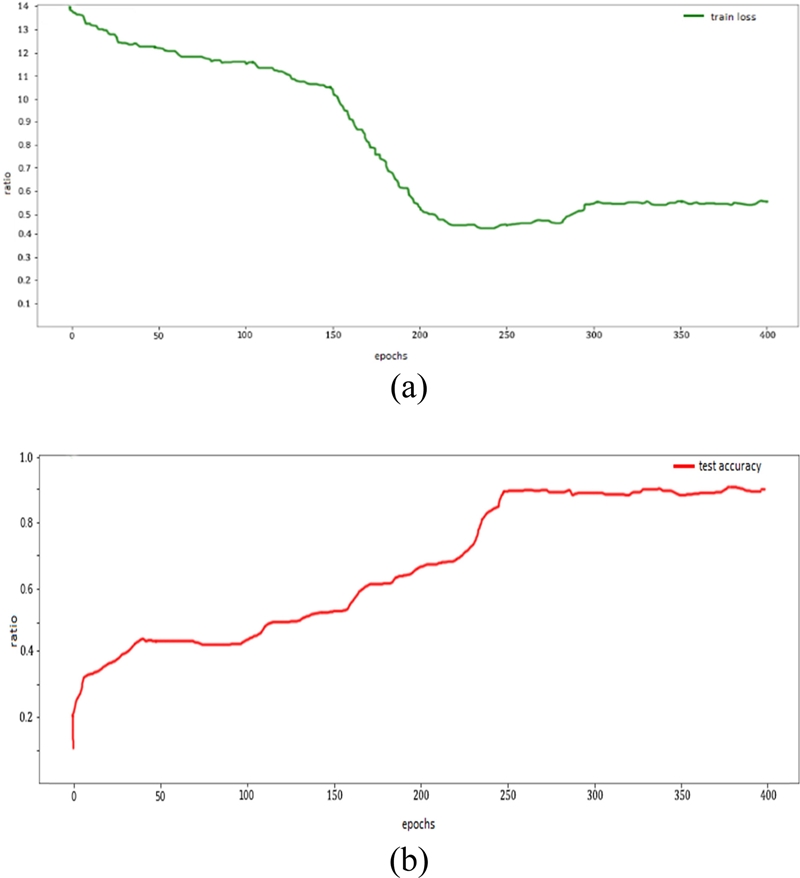
The accuracy of the MLP, CNN, and SNN are analyzed in Figures 13, 14, and 15, respectively. The CNN achieved the highest accuracy of approximately 95% and the loss decreased from 1.6 to 0.2. The SNN achieved an accuracy of 90%, with a decrease in error rate from 1.4 to 0.6 with 400 epochs.
4.2 Power Consumption
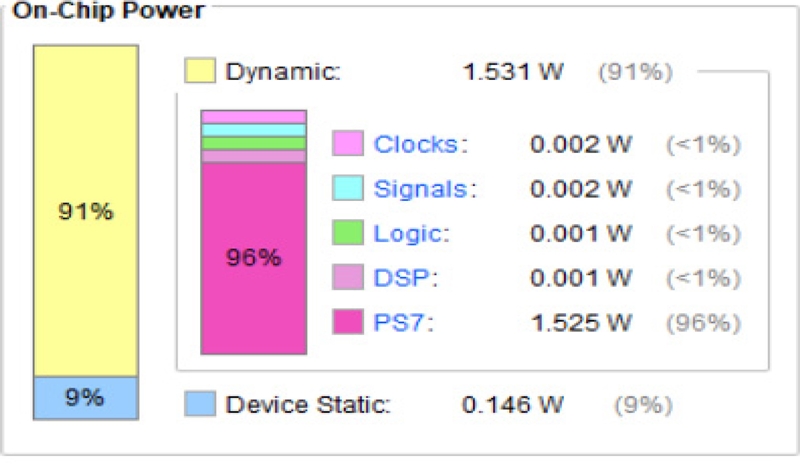
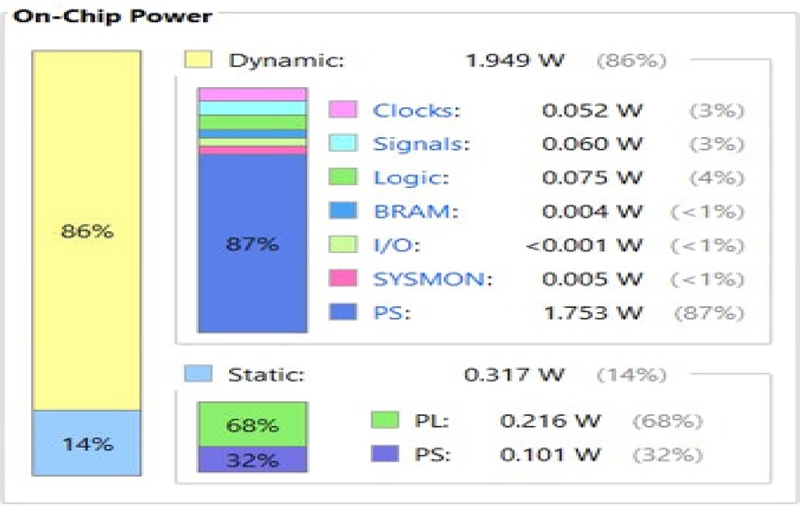
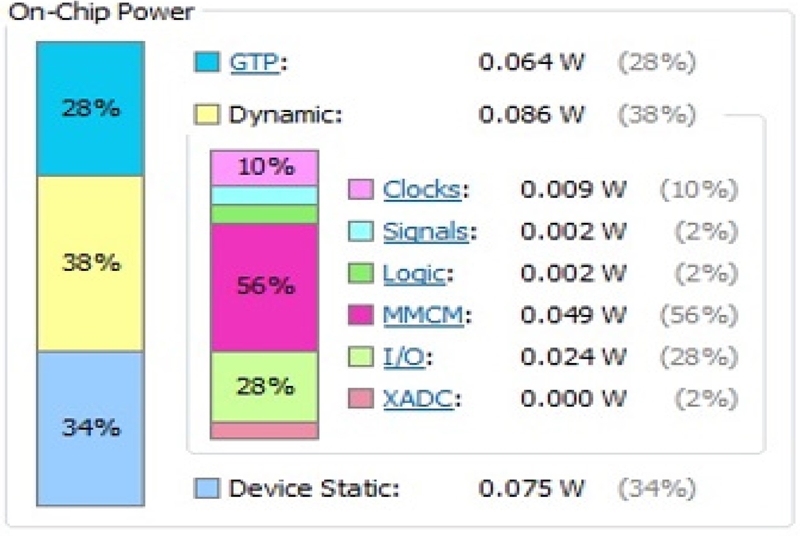
The power consumption of the three AI algorithms is shown in Figures 16, 17, and 18. The SNN outperforms the other two AI algorithms in terms of power. Table 2 shows the comparison of the three ANNs based on accuracy, time, and power. Among the three ANNs, the SNNs consumed the least power (0.226 W).
5. CONCLUSIONS
In the simulation results, as the CNN consumed more time for training, i.e., almost 1 h, it was considered as the most power consuming algorithm as the hidden layers were dense compared to other networks, which resulted in a bulky operation. The computation amount increased with the number of hidden layers in the network.
In addition, the SNN performed computations with only two hidden layers and could yield a maximum accuracy of 90%. The advantage of using the SNN is that it propagates only when the spike interval of a neuron reaches a certain threshold. However, the MLP and CNN propagate oppositely. Therefore, less computational source and time were required by the SNN for the ECG classification system. The SNN was the most efficient ECG classification algorithm, which classified all four ECG arrhythmias with 90 % accuracy.
In the future, ECG arrhythmias will be classified more accurately, and the power consumption will be reduced to less than 1 uW. Moreover, it is believed that SNNs may easily enable low-power hardware evaluation.
References
- R. Lozano, M. Naghavi, K. Foreman, S. Lim, K. Shibuya, V. Aboyans, J. Abraham, T. Adair, R. Aggarwal, S. Y. Ahn, et al., “Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010”, The Lancet, Vol. 380, No. 9859, pp. 2095-2128, 2013.
-
M. Mitra and R. K Samanta, “Cardiac Arrhythmia Classification Using Neural Networks with Selected Features: first international conference on Computational Intelligence: Modeling Techniques and Applications (CIMTA)”, pp. 76-84, 2013.
[https://doi.org/10.1016/j.protcy.2013.12.339]

- T. Habte, T. Saleh, H. Mohammad, B, & Ismail, M., “Ultra Low Power ECG Processing System for IoT Devices: Analog Circuits and Signal Processing”, pp. 1-14, 2019.
-
T. E. Taha, A. E. Sayed, and S. Rafat, “A survey on Classification of ECG Signal Study”, Vol. 6, No. 5, pp. 11-16, 2016.
[https://doi.org/10.5120/cae2016652467]

- C. V. Maggio, A., Bonomini, M., Laciar, E., and Arini, P., “Quantification of Ventricular Repolarization Dispersion Using Digital Processing of the Surface ECG” , 2012.
-
M. Aqil,A. Jbari, and A. Bourouhou, “ECG signal denoising by Discrete Wavelet Transform”, Vol. 13, No. 09, pp. 51-68, 2017.
[https://doi.org/10.3991/ijoe.v13i09.7159]

- https://blog.octo.com/en/time-series-features-extraction-usingfourier-and-wavelet-transforms-on-ecg-data, /, (retrieved on Nov. 23, 2019).
- Bultheel Bull. Belg. B. Yu, M. Vetterli, “Chapter 4. Data Processing and Analysis, Math Wavelet Transform” IEEE Pub., pp. 35-42, 1995.
- M. Sanaullah, “A review of higher order statistics and spectra in communication systems”, Vol. 13, No. 4, pp. 31-50, 2013.
- https://towardsdatascience.com/a-one-stop-shop-for-principal-component-analysis-5582fb7e0a9c, (retrieved on Apr 18, 2017).
-
E. D. Ubeyli, “Combining recurrent neural networks with eigenvector methods for classification of ECG beats. Digital Signal Processing”, Vol. 19, Issue 2, pp. 320-329, 2009.
[https://doi.org/10.1016/j.dsp.2008.09.002]

-
J. Park, K. Lee, K. Kang, “Arrhythmia detection from heartbeat using the k-nearest neighbor classifier.” IEEE International Conference on Bioinformatics and Biomedicine, pp 15-22 (2013).
[https://doi.org/10.1109/BIBM.2013.6732594]

-
S. Kiranyaz, T. Ince, M. Gabbouj, “Real-time patient-specific ECG classification by 1-D convolutional neural networks”, IEEE Transactions on Biomedical Engineering, Vol. 63, No. 3, pp.664-675, 2016.
[https://doi.org/10.1109/TBME.2015.2468589]

- T. J. Jun, H. M. Nguyen, D. Kang, D. Kim, D. Kim, and Y.H. Kim, “ECG arrhythmia classification using a 2-D convolutional neural network”, 18 Apr 2018.
-
S. Savalia and V. Emamian, “Cardiac Classification by Multi-Layer Perceptron and Convolutional Neural Networks”, Vol. 5, No. 35, pp. 1-12, 2018.
[https://doi.org/10.3390/bioengineering5020035]

-
A. Das, P. Pradhapan, W. Groenendaal, P. Adiraju, R. T. Rajan, F. Catthoor, S. Schaafsma, J.L, Krichmar, N.Dutt, and C. Van Hoot, “Unsupervised heart-rate estimation in wearable with liquid states and a probabilistic readout”, Neural Networks. Vol.99, pp.134-147, 2018.
[https://doi.org/10.1016/j.neunet.2017.12.015]

-
A. Amirshahi and M. Hashemi, “Ultra Low-Power and Real-time ECG classification Based on STDP and R-STDP Neural networks for Wearable Devices”, IEEE Transactions on Biomedical Circuits and Systems (TBioCAS), Vol. 13, Issue. 6, pp. 1483-1493, 2019.
[https://doi.org/10.1109/TBCAS.2019.2948920]

- K. Guo, J. Yu, Y. Wang, and H. Yang, “A survey of FPGA Based Neural Network Accelerator”, ACM Transactions on Reconfigurable Technology and Systems, Vol. 9, No. 4, Article 11, Dec 2017.
-
C. V. Nguyen, T. Le Quang, T. N. Vu, H. Le Thi, K. N. Van, T. H. Trong, K. Ishibashi, “A non-contact infection screening system using medical radar and Linux-embedded FPGA: Implementation and preliminary validation.”, Informatics in Medicine Unlocked, Vol.16, pp. 1-9, 2019.
[https://doi.org/10.1016/j.imu.2019.100225]