Immunoaffinity Characteristics of Exosomes from Breast Cancer Cells Using Surface Plasmon Resonance Spectroscopy
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License(http://creativecommons.org/licenses/bync/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Exosomes, known as nanoscale extracellular vesicles in the range of 30–150 nm, are known to contain clinically significant information. However, there is still insufficient information on exosomal membrane proteins for cancer diagnosis. In this work, we investigated the characteristics of the membrane proteins of exosomes shed by cultured breast cancer cell lines using a surface plasmon resonance (SPR) spectroscopy and pre-activated alkanethiols modified sensor chips. The antibodies of breast cancer biomarkers such as MCU-16, EpCAM, CD24, ErbB2, and CA19-9 were immobilized on the pre-activated alkanethiols surfaces without any activation steps. The purified exosomes were loaded onto each antibody surface. The affinity rank of the antibody surfaces was decided by the relative capture efficiency factors for the exosomes. In addition, an antibody with a relative capture efficiency close to 100% was tested with exosome concentration levels of 104/μl, 105/μl, and 106/μl for quantitative analysis.
Keywords:
Exosome, Breast cancer, Surface plasmon resonance, Relative capture efficiency factor, Antibody1. INTRODUCTION
The exosomes secreted by various cells are small extracellular vesicles with a size of 30–150 nm and are found in body fluids such as blood, urine, saliva, and breast milk. Because exosomes contain proteins and RNAs related to the cells of origin, they are known to represent the characteristics and states of the cells. In particular, exosomes play a key role in intercellular communication, and thereby intensive research on their physiological and immunological roles has been undergoing in order to utilize them as biomarkers for diagnostics and in therapeutics by controlling the secretion of exosomes from cells [1-8]. Exosomes have been reportedly found in body fluids of healthy individuals; however, their concentrations change in a diseased state [9,10]. Breast cancer is the most commonly diagnosed cancer among women, and it is a leading cause of cancer mortality in women worldwide [11,12]. MCU-16 [13], EpCAM [14], CD24 [15], ErbB2 [16], and CA19-9 [17] are the known biomarkers of breast cancer. As the importance of exosomes becomes significant, efficient techniques for exosome concentration measurement are highly required.
Nowadays, one of the popular analytical techniques to determine the concentration of biomolecules is the use of surface plasmon resonance (SPR) sensors because these optic-based sensors are highly sensitive in the vicinity of a metal (conventionally gold (Au)) chip surface. [18, 19]. Resonance occurs between incident light and surface plasmon, resulting in the minimum reflected light intensity. This resonance exhibits a sharp dip in the reflectance curve, and the angle at which the reflectivity is the minimum is called as the SPR angle. The SPR sensing is based on the variation in reflectivity that resulted from refractive index changes in the vicinity of the metal surface. In general, the change in refractive index due to biomolecular interactions can be translated as the change in the SPR angle [18, 19]. The use of this powerful analytical technique includes fundamental biochemical research such as protein-protein and protein-DNA interactions, and other applications in diagnostics, drug discovery, environmental monitoring, and food safety [20-24]. The significant advantages of the SPR sensors are label-free detection, direct measurement, and versatility owing to the use of a variety of materials [18, 19]. Additionally, the effective sensing distance of SPR sensors is up to 200 nm by surface, and their sensitivity is maximized when the analytes exist within this effective sensing distance. Because exosomes have diameters of less than 200 nm, using an SPR sensor is one of the most appropriate methods to detect them.
In this study, an SPR technique for exosome characterization was investigated using the antibodies of breast cancer biomarkers. After the functionalization of sensor surfaces with the related antibodies (MCU-16, EpCAM, CD24, ErbB2, and CA19-9), purified exosomes isolated from MCF-7, i.e., breast cancer cells, were injected into the flow cell of an SPR system for analysis. Responses of exosomes to the antibody-functionalized surfaces were investigated, and a relative capture efficiency factor is introduced to understand the exosome capture efficiency of the corresponding antibody. These measurements and the introduced factor would help find the efficient methods to diagnose the breast cancer using exosomes.
2. EXPERIMENTAL
2.1 Cell culture
An MCF-7 human breast cancer cell was cultured in Dulbecco’s minimal essential medium (DMEM; Biowest) supplemented with 10% (v/v) fetal bovine serum (FBS, Biowest) and 1% (v/v) penicillin and streptomycin (Life Technologies) in a humidified 5% CO2 atmosphere at 37 °C. After the cells were grown to 70% confluency for three days in the media containing 10% FBS, they were washed twice with phosphate buffered saline (PBS) and incubated in 10% exosome-free FBS DMEM for an additional two days.
2.2 Isolation of exosomes
In this study, the exosomes were isolated using a precipitation method that used a commercially available reagent (ExoQuick-TCTM: System Biosciences) according to the manufacturer’s instructions after the cell culture. The media were centrifuged at 3000 × g for 15 min to remove cells and their debris. The supernatant was mixed with ExoQuick- TCTM solution. The mixture remained upright at 4 °C overnight for incubation. Subsequently, the mixture was centrifuged at 1500 × g for 30 min. The pellets containing the exosomes were resuspended in PBS.
2.3 SPR measurements
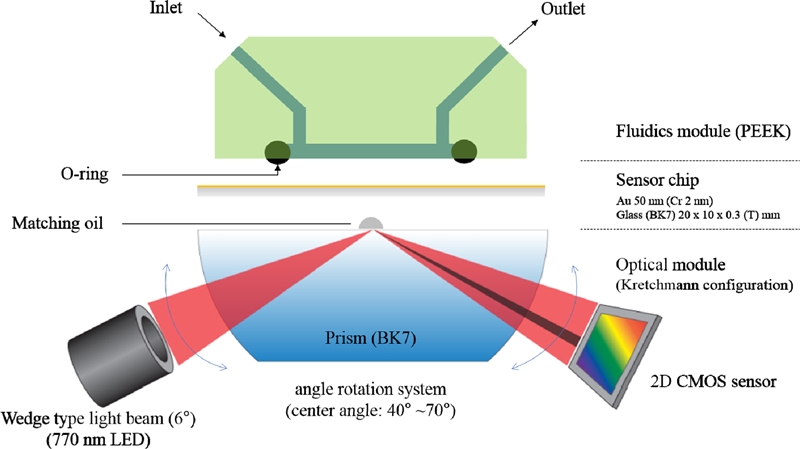
SPR measurements were performed on an iMstart400N instrument (Innotoolsfactory, Rep. of Korea) at a temperature of 22 °C at a flow rate of 30 μl/min in a rectangular flow cell. The SPR instrument was based on the Kretschman configuration with a light-emitting diode (LED) as the light source that had a peak wavelength of 770 nm (OpNext Inc., Japan) with band pass filter (± 10 nm) and a 2-dimensional complementary-metaloxide semiconductor (2D-CMOS: 1280×1024 pixels, IDS Co., Germany) image sensor in combination with a BK-7 prism (Fig. 1). The SPR chip is a stack of 48 nm of Au and 2 nm of chromium (Cr) as an adhesion layer. The refractive index of the Au at 770 nm is 0.14430 + i4.6583. The angle of the wedge-shaped beam was 6°. The incident angle was optimized so that the SPR occurred in the middle of channels to measure the variations. The measurements were carried out in angular interrogation mode. The minimum detectable SPR angle shift is 0.0001°.
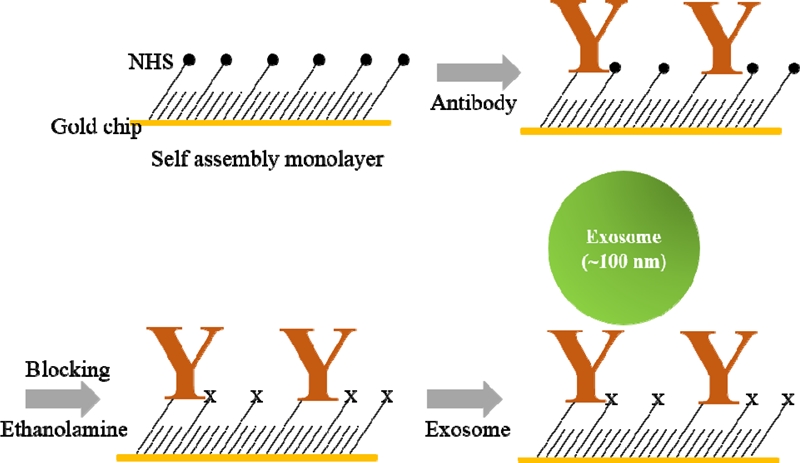
A self-assembled monolayer (SAM) based on 5 mM of 11-Mercapto-1-undecanol with a disulfide bond combined with N– hydroxysulfosuccinimide (NHS) was acquired from Innotoolsfactory and was then formed on the Au surfaces of SPR chips (Innotoolsfactory, Rep. of Korea) by overnight immersion in SAM-dissolved chloroform (Sigma, USA). The antibodies (10 μg/ ml in 1 × PBS) were injected into the flow cell of the SPR sensor system for immobilization of the antibodies on the NHS-SAM functionalized SPR chip, and the unbound antibodies were removed by flushing with a 1 × PBS solution. 1 M ethanolamine (in DI-water) was used to block non-specific bindings. Finally, sample solutions containing exosomes were sequentially injected over the antibody-immobilized Au SPR chips. Fig. 2 shows schemes of orders for the functionalization of the SPR sensor surface. 1 × PBS and ethanolamine were purchased from Sigma (USA), and the antibodies (Anti-EpCAM, Anti-CD24, Anti-MUC16, Anti-CA19-9, Anti-ErbB2, IgG) were acquired from Abcam (USA).
3. RESULTS AND DISCUSSIONS
Fig. 3 shows a typical SPR sensorgram. After SAM formation on the SPR chip surface, the functionalized surface was sequentially exposed to the slowly-flowing antibodies of breast cancer markers and 1 M of ethanolamine followed by the isolated exosomes. In Fig. 3, the SPR signal magnitude difference from the baseline indicates the response of an antibody to the SAM surface after injection of the antibody and flushing of any unbound antibody. When the ethanolamine was injected into the SPR system, the signal initially elevated and then descended after the PBS buffer solution was injected again. This is because the refractive index of the ethanolamine solution diluted by DI water is lower than that of the PBS solution. In this case, however, the magnitude of the SPR signal before and after the injection of ethanolamine did not change significantly. This is because, unlike the antibody binding process, the ethanolamine treatment process did not change the mass on the surface of the sensor chip. Injection of the exosome solutions increased the signal and produced a stable signal after the PBS buffer solution was injected for washing as shown in Fig. 3. Therefore, we can conclude that a functionalized SPR sensor surface can capture exosomes efficiently.
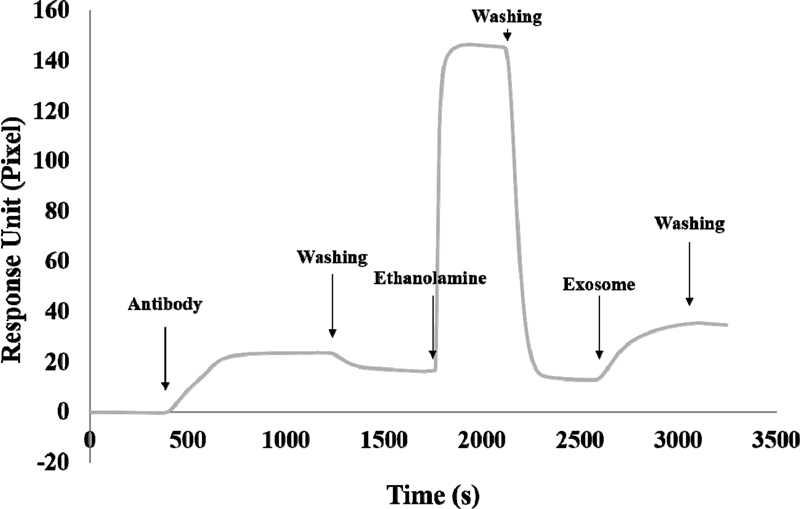
A typical SPR sensorgram of the SPR sensor chip by sequence injection of an antibody, ethanolamine, and exosome.
Fig. 4(a) shows histograms representing the reaction of the five antibodies to the SAM functionalized surface and the capture of exosomes from the MCF-7 cell by the five different types of immobilized antibodies: anti-EpCAM, anti-CD24, anti-MUC16, anti-CA19-9, and anti-ErbB2. The values were the mean responses of two identical experiments, and the error bars represent the standard deviation from two replicates. In Fig. 4(a), among the five antibodies immobilized by the SAM, anti-MUC16 was found to have the highest exosome capture capability. From the response histogram in Fig. 4(a), the largest amount of anti-ErbB2 was immobilized on the SAM-functionalized surface. However, the quantity of captured exosomes was not the largest among the five antibodies. Compared with anti-ErbB2, anti-MUC16 showed a lower response signal for immobilization on the SAM-functionalized surface. Here, a relative capture efficiency factor is introduced to understand the exosome capture efficiency (capability) of the antibody. The relative capture efficiency factor is defined as the ratio of the exosome’s response to the immobilized antibody to the antibody’s response to the SAM-functionalized surface chip. The relative capture efficiency factor indicates the quantity of exosomes that can be captured by a unit antibody response. The relative capture efficiency factors for the five antibodies are given in Fig. 4(b). The relative capture efficiency factor for anti-MUC16 was the highest, which implies that anti-MUC16 can efficiently capture a large number of exosomes from breast cancer cells. The high response that anti-MUC16 exhibited to the exosomes from MCF-7 cells may imply that the marker protein on the surface of the exosome expressed MUC16 in abundance. MCU16 is not a specific biomarker of breast cancer as its elevation can be found in other types of tumors such as ovarian, endometrial, pancreatic, colon, and lung cancers [13]. ErbB2 is relatively more specific to breast cancer [16, 25], and it also exhibited a high response to exosomes from the breast cancer cells. Thus, simultaneous detection of exosomes using both anti-MUC16 and anti-ErbB2 would give specific information.

(a) Responses of five antibodies to the SAM-functionalized surface and exosomes from MCF-7 cell to the five immobilized antibodies: anti-EpCAM, anti-CD24, anti-MUC16, anti-CA19-9, and anti-ErbB2. (b) The relative capture efficiency factors for the five antibodies.
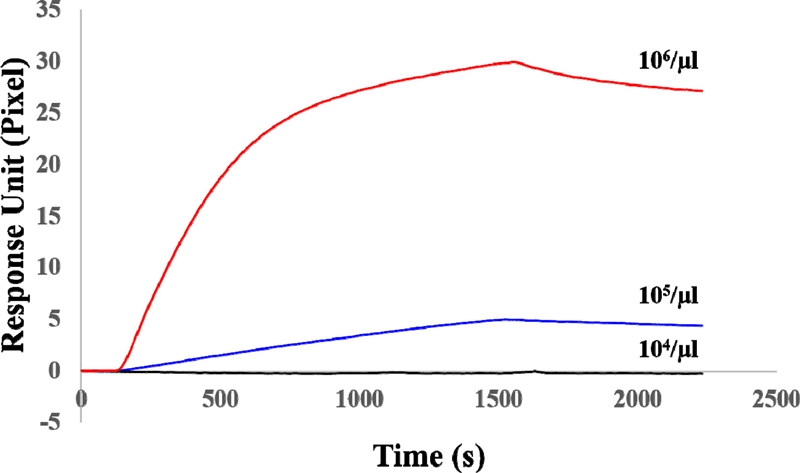
Among the five antibodies, anti-EpCAM was selected to investigate what concentrations of the exosome can cause this SPR system to respond as its relative capture efficiency factor was very close to 100%. Fig. 5 shows a sensorgram of the response of the anti-EpCAM-functionalized SPR chip to the exosomes with concentrations of 104/μl, 105/μl, and 106/μl. The experimental procedure was the same as the one carried out for the result of the sensorgram in Fig. 3. The experiment was performed by sequence injection of anti-EpCAM for the antibody immobilization, ethanolamine to prevent non-specific reaction, and finally the exosomes of various concentrations. The vertical axis of Fig. 5 represents the amount of change in the CMOS pixel of the SPR angle after exosome injection. As shown in Fig. 5, when the concentration of exosomes was 104/μl, no significant signal variation occurred. However, the reaction of the SPR chip was observed to begin responding when the concentration of exosomes was 105/μl or more. This suggests that to quantify exosomes, at least more than 105/μl exosomes are required to respond in this SPR system for the anti-EpCAM-functionalized SPR chip. The gold standard method for extracting exosomes is the ultracentrifuge method. The method provides high-purity exosomes but has the disadvantage of a low yield. Generally, the amount obtained when extracting exosomes from human serum using ultracentrifuge is 106/μl [26]. The minimum concentration of 105/μl observed in this experiment was ten times less than that obtained using the ultracentrifuge method; therefore, this approach provides sufficient amounts for exosome analysis.
4. CONCLUSIONS
An SPR platform based on the Kretschman configuration was used to investigate immunoaffinity characteristics of the exosomes secreted by an MCF-7 human breast cancer by functionalizing the sensor surfaces with anti-MCU-16, anti-EpCAM, anti-CD24, anti-ErbB2, and anti-CA19-9 antibodies. The relative capture efficiency factor was adopted to understand the exosome capture efficiency of the antibodies. The relative capture efficiency factor for anti-MUC16 was the highest among the above-mentioned antibodies, which implies that anti-MUC16 can efficiently capture exosomes from the breast cancer cells. The anti-EpCAM whose relative capture efficiency factor was very close to 100% was utilized to investigate the reaction concentrations of the exosome for this SPR system, and the functionalized SPR chip began to respond when the concentration of the exosomes was 105/μl. Simultaneous detection with specific immunosensor surfaces will pave the way for cancer diagnosis and personalized medicine using less-invasive methods.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government, MSIP (NRF-2016R1A5A1010148), and Individual Basic Science & Engineering Research Program (NRF- 2018R1D1A1B07048926) of the National Research Foundation of Korea, funded by the Ministry of Education.
References
-
C. Lawson, J. M. Vicencio, D. M. Yellon, and S. M. Davidson, “Microvesicles and exosomes: new players in metabolic and cardiovascular disease”, J. Endocrinol., Vol. 288, pp. R57-R71, 2016.
[https://doi.org/10.1530/JOE-15-0201]

-
S. H. Jalalian, M. Ramezani, S. A. Jalalian, K. Abnous, and S. M. Taghdisi, “Exosomes, new biomarkers in early cancer detection”, Anal. Biochem., Vol. 571, pp. 1-13, 2019.
[https://doi.org/10.1016/j.ab.2019.02.013]

-
L. Console, M. Scalise, and C. Indiveri, “Exosomes in inflammation and role as biomarkers”, Clin. Chim. Acta, Vol. 488, pp. 165-171, 2019.
[https://doi.org/10.1016/j.cca.2018.11.009]

-
M. R. Fernando, C. Jiang, G. D. Krzyzanowski, and W. L. Ryan, “New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes,” PLoS ONE, Vol. 12, pp. e0183915(1)-e0183915(15), 2017.
[https://doi.org/10.1371/journal.pone.0183915]

-
T. Pisitkun, R. F. Shen, and M. A. Knepper, “Identification and proteomic profiling of exosomes in human urine”, Proc. Natl. Acad. Sci. U.S.A., Vol. 101, pp. 13368-13373, 2004.
[https://doi.org/10.1073/pnas.0403453101]

-
V. Palanisamy, S. Sharma, A. Deshpande, H. Zhou, J. Gimzewski, and D. T. Wong, “Nanostructural and transcriptomic analyses of human saliva derived exosomes”, PLoS ONE, Vol. 5, pp. e8577(1)-e8577(11), 2010.
[https://doi.org/10.1371/journal.pone.0008577]

-
C. Admyre, S. M. Johansson, K. R. Qazi, J.-J. Filén, R. Lahesmaa, M. Norman, E. P. A. Neve, A. Scheynius, and S. Gabrielsson, “Exosomes with Immune Modulatory Features Are Present in Human Breast Milk”, J. Immunol., Vol. 179, pp. 1969−1978, 2007.
[https://doi.org/10.4049/jimmunol.179.3.1969]

-
S. W. Ferguson and J, Nguyen, “Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity”, J. Control. Release, Vol. 228, pp. 179-190, 2016.
[https://doi.org/10.1016/j.jconrel.2016.02.037]

-
C. Beyer and D. S. Pisetsky, “The role of microparticles in the pathogenesis of rheumatic diseases”, Nat. Rev. Rheumatol., Vol. 6, pp. 21−29, 2010.
[https://doi.org/10.1038/nrrheum.2009.229]

-
D. L. M. Rupert, C. Lässer, M. Eldh, S. Block, V. P. Zhdanov, J. O. Lotvall, M. Bally, and F. Höök, “Determination of Exosome Concentration in Solution Using Surface Plasmon Resonance Spectroscopy”, Anal. Chem., Vol. 86, pp. 5929–5936, 2014.
[https://doi.org/10.1021/ac500931f]

-
L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent, and A. Jemal, “Global cancer statistics, 2012”, CA. Cancer J. Clin., Vol. 65, pp. 87-108, 2015.
[https://doi.org/10.3322/caac.21262]

-
R. Siegel, D. Naishadham, and A. Jemal, “Cancer statistics, 2012”, CA. Cancer J. Clin., Vol. 62, pp. 10-29, 2012.
[https://doi.org/10.3322/caac.20138]

-
S. Reinartz, S. Failer, T. Schuell, and U. Wagner, “CA125 (MUC16) gene silencing suppresses growth properties of ovarian and breast cancer cells”, Eur. J. Cancer, Vol. 48, pp. 1558-1569, 2012.
[https://doi.org/10.1016/j.ejca.2011.07.004]

-
N. M. R. Abd El-Maqsoud and D. M. Abd El-Rehim, “Clinicopathologic Implications of EpCAM and Sox2 Expression in Breast Cancer”, Clin. Breast Cancer, Vol. 14, pp. e1-e9, 2014.
[https://doi.org/10.1016/j.clbc.2013.09.006]

-
K. Suyama, H. Onishi, A. Imaizumi, K. Shinkai, M. Umebayashi, M. Kubo, Y. Mizuuchi, Y. Oda, M. Tanaka, M. Nakamura, and M. Katano, “CD24 suppresses malignant phenotype by downregulation of SHH transcription through STAT1 inhibition in breast cancer cells”, Cancer Lett., Vol. 374, pp. 44-53, 2016.
[https://doi.org/10.1016/j.canlet.2015.12.013]

-
U. Eletxigerra, J. Martinez-Perdiguero, S. Merino, R. Barderas, R. M. Torrente-Rodríguez, R. Villalonga, J. M. Pingarrón, and S. Campuzano, “Amperometric magnetoimmunosensor for ErbB2 breast cancer biomarker determination in human serum, cell lysates and intact breast cancer cells”, Biosens. Bioelectron., Vol. 70, pp. 34-41, 2015.
[https://doi.org/10.1016/j.bios.2015.03.017]

-
W. Wang, X. Xu, B. Tian, Y. Wang, L. Du, T. Sun, Y. Shi, X. Zhao, and J. Jing, “The diagnostic value of serum tumor markers CEA. CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer”, Clin. Chim. Acta, Vol. 470, pp. 51-55, 2017.
[https://doi.org/10.1016/j.cca.2017.04.023]

-
H. Šípová and J. Homola, “Surface plasmon resonance sensing of nucleic acids: A review”, Anal. Chim. Acta, Vol. 773, pp. 9-23, 2013.
[https://doi.org/10.1016/j.aca.2012.12.040]

-
C. Liu, F. Hu, W. Yang, J. Xu, and Y. A. Chen, “Critical review of advances in surface plasmon resonance imaging sensitivity”, Trends Anal. Chem., Vol. 97, pp. 354-362, 2017.
[https://doi.org/10.1016/j.trac.2017.10.001]

-
C. Lertvachirapaiboon, A. Baba, S. Ekgasit, K. Shinbo, K. Kato, and F. Kaneko, “Transmission surface plasmon resonance techniques and their potential biosensor applications”, Biosens. Bioelectron., Vol. 99, pp. 399-415, 2018.
[https://doi.org/10.1016/j.bios.2017.07.069]

-
C. G. Zhang, S. J. Chang, K. Settu, C. J. Chen, and J. T. Liu, “High-sensitivity glycated hemoglobin (HbA1c) aptasensor in rapid-prototyping surface plasmon resonance”, Sens. Actuators B, Vol. 279, pp. 267-273, 2019.
[https://doi.org/10.1016/j.snb.2018.09.077]

-
S. G. Patching, “Surface plasmon resonance spectroscopy for characterisation of membrane protein–ligand interactions and its potential for drug discovery,” Biochim. Biophys. Acta Biomembr., Vol. 1838, pp. 43-55, 2014.
[https://doi.org/10.1016/j.bbamem.2013.04.028]

-
M. Mahmoudpour, J. E. N. Dolatabadi, M. Torbati, and A. Homayouni-Rad, “Nanomaterials based surface plasmon resonance signal enhancement for detection of environmental pollutions”, Biosens. Bioelectron., Vol. 127, pp. 72-84, 2019.
[https://doi.org/10.1016/j.bios.2018.12.023]

-
B. J. Yakes, J. Buijs, C. T. Elliott, and K. Campbell, “Surface plasmon resonance biosensing: Approaches for screening and characterizing antibodies for food diagnostics”, Talanta, Vol. 156-157, pp. 55-63, 2016.
[https://doi.org/10.1016/j.talanta.2016.05.008]

-
O. P. Kallioniemi, A. Kallioniemi, W. Kurisu, A. Thor, L. C. Chen, H. S. Smith, F. M. Waldman, D. Pinkel, and J. W. Gray, “ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization”, Proc. Natl. Acad. Sci. U.S.A., Vol. 89, pp. 5321–5325, 1992.
[https://doi.org/10.1073/pnas.89.12.5321]

-
I. Helwa, J. Cai, M. D. Drewry, A. Zimmerman, M. B. Dinkins, M. L. Khaled, M. Seremwe, W. Michael Dismuke, E. Bieberich, W. D. Stamer, M. W. Hamrick, and Y. Liu, “Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents”, PLoS ONE, Vol. 12, pp. e0170628(1)-e0170628(22), 2017.
[https://doi.org/10.1371/journal.pone.0170628]